using Distributions, Zygote, Plots, StatsPlots, RandomCensored Data
https://juliastats.org/Distributions.jl/stable/censored/
Random.seed!(123)
line = collect(-1:0.01:3)
censoring = 1.5
actual = rand(Normal(1,0.5),500)
observed = min.(censoring,actual)
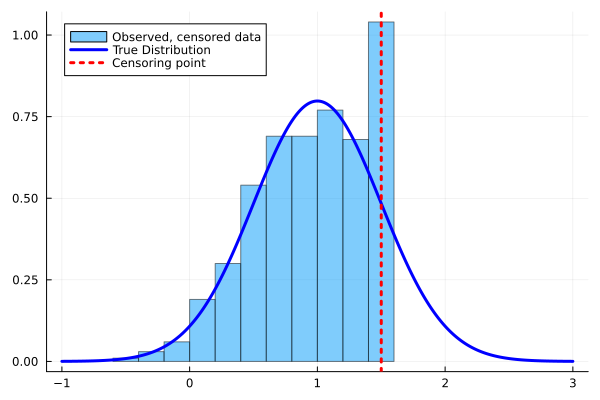
histogram(observed,
normalize=:true,
label = "Observed, censored data",
alpha=0.5,
legend=:topleft,
fmt=:png
)
plot!(line,
pdf.(Normal(1,0.5),line),
lw=3,
c=:blue,
label = "True Distribution"
)
vline!([censoring],
lw=3, c="red",
ls=:dot,
label="Censoring point"
)
mean_full = mean(observed)
std_full = std(observed)
mean_red = mean(observed[observed .< censoring])
std_red = std(observed[observed .< censoring])
ps = [0.,0.]
for i in 1:50
grads = Zygote.gradient(p->-mean(logpdf.(censored.(Normal(p[1],exp(p[2])),-Inf,ones(length(observed)).*censoring),observed)),ps)[1]
ps .-= 0.1 .* grads
end
mean_model = ps[1]
std_model = exp(ps[2])
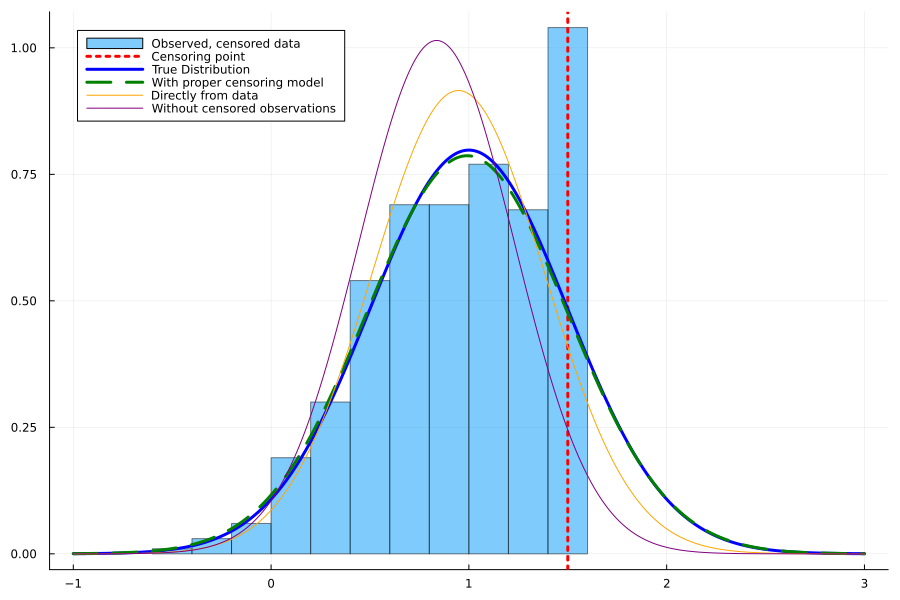
histogram(observed,
normalize=:true,
label = "Observed, censored data",
alpha=0.5,
legend=:topleft,
fmt=:png,
size=(900,600)
)
vline!([censoring],
lw=3, c="red",
ls=:dot,
label="Censoring point"
)
plot!(line,
pdf.(Normal(1,0.5),line),
lw=3,
c=:blue,
label = "True Distribution"
)
plot!(line,
pdf.(Normal(mean_model,std_model),line),
lw=3,
c=:green,
s=:dash,
label = "With proper censoring model"
)
plot!(line,
pdf.(Normal(mean_full,std_full),line),
lw=1,
c=:orange,
label = "Directly from data"
)
plot!(line,
pdf.(Normal(mean_red,std_red),line),
lw=1,
c=:purple,
label = "Without censored observations"
)