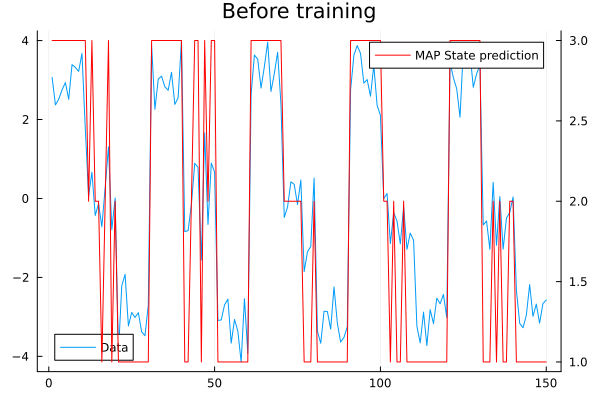
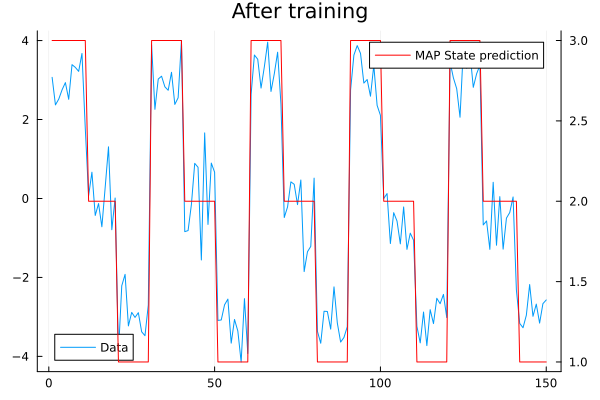
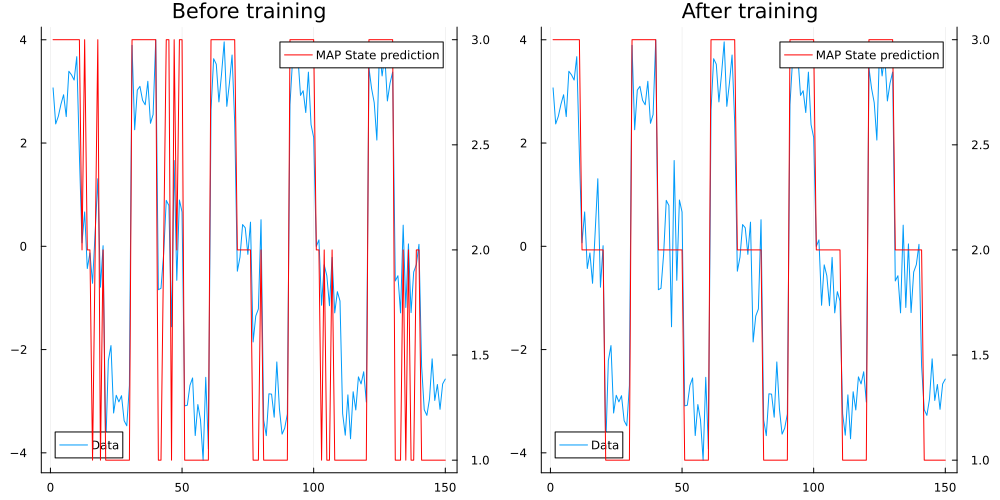
mutable struct HMM
μ
σ
P
end
Flux.@functor HMM (μ, σ)
function HMM(states)
μ = collect(range(-1,1, length=states))
σ = zeros(states)
P = softmax(Matrix(Diagonal(ones(states))),dims=2)
return HMM(μ,σ,P)
end
function filter(m::HMM, y, p_t)
μ = m.μ
σ = exp.(m.σ)
P = m.P
y_t = y[1]
dists_t = Normal.(μ,σ)
pdfs = pdf.(dists_t, y_t)
sumdist = (p_t.*pdfs)
p_tt = sumdist./sum(sumdist)
p_tp1 = P*p_tt
if length(y)>1
dists_tp1, p_t, p_ttp1 = filter(m,y[2:end],p_tp1)
return vcat(dists_t, dists_tp1), hcat(p_t,p_tp1), hcat(p_tt, p_ttp1)
else
return dists_t, p_t, p_tt
end
end
function forward(m::HMM, y, α_tm1)
#α(q_0) = p(q_0)p(y_0|q_0)
μ = m.μ
σ = exp.(m.σ)
P = m.P
y_t = y[1]
qsum = P*α_tm1
dists = Normal.(μ,σ)
pdfs = pdf.(dists,y_t)
α_t = pdfs.*qsum
if length(y)>1
α_tp1 = forward(m,y[2:end],α_t)
return hcat(α_t, α_tp1)
else
return α_t
end
end
function backward(m::HMM, y, β_tp1)
#β_(q_{T-1}) = \sum_{q_T}p(y_T|q_T)p(q_T|q_{T-1})
μ = m.μ
σ = exp.(m.σ)
P = softmax(m.P, dims=2)
y_t = y[end]
dists = Normal.(μ,σ)
pdfs = pdf.(dists,y_t)
β_t = sum(transpose(P).*β_tp1.*pdfs,dims=2)
if length(y)>1
β_tm1 = backward(m, y[1:end-1], β_tp1)
return hcat(β_tm1, β_t)
else
return β_t
end
end
function likelihood(m::HMM,y,sps)
μ = m.μ
σ = exp.(m.σ)
dists = Normal.(μ,σ)
return mean(map(i->sum(sps[i].*logpdf.(dists,y[i])),1:length(y)))
end
function EM(m::HMM, y, p_0, n_iter = 50)
for i in 1:n_iter
αβ = backward(m,y,p_0).*forward(m,y,p_0)
state_probs = αβ./sum(αβ,dims=1)
sps = Flux.unstack(state_probs,dims=2)
ps, f = Flux.destructure(m)
for _ in 1:50
grads = ForwardDiff.gradient(x -> -likelihood(f(x),y,sps), ps)
ps .-= 0.001.*grads
end
newm = f(ps)
m.μ = newm.μ
m.σ = newm.σ
Ps = Matrix(transpose(hcat([sum(state_probs[i:i, 1:end-1].*state_probs[:, 2:end],dims=2)[:] for i in 1:length(p_0)]...)))
Ps./=sum(Ps,dims=1)
m.P = Ps
if i%50==0
println(-likelihood(m,y,sps))
end
end
end